Helix Synth is a three-phase powerhouse, mixing superior AI with organic information to deal with protein construction prediction.
Helix Synth begins with huge datasets from sources like UniProt, DSSP, and the RCSB Protein Knowledge Financial institution (PDB). These datasets label proteins into three secondary construction sorts: H (Helix), E (Beta Sheet), and C (Coil). The info is preprocessed utilizing:
- Characteristic Extraction: Sequences are encoded with one-hot encoding and pretrained embeddings like ProtBERT, TAPE, and ESM2.
- Tensor Prep: NumPy and Pandas deal with information for GPU-friendly batching.
- Coaching: The mannequin trains on Kaggle T4 GPUs with CUDA, utilizing tips like batch processing and torch.cuda.empty_cache() to optimise efficiency. Coaching stops early after 30 epochs to keep away from overfitting.
The structure is a rigorously crafted ensemble:
- CNNs seize native patterns in protein sequences.
- BiLSTM fashions long-range dependencies, essential for understanding complicated folds.
- Absolutely related layers and softmax classify buildings with confidence scores.
- Adam Optimiser and Cross-Entropy Loss guarantee quick, correct studying.
The outcome? An general accuracy of 71.01%, with particular accuracies of 76.21% (helix), 63.26% (beta sheet), and 70.92% (coil).
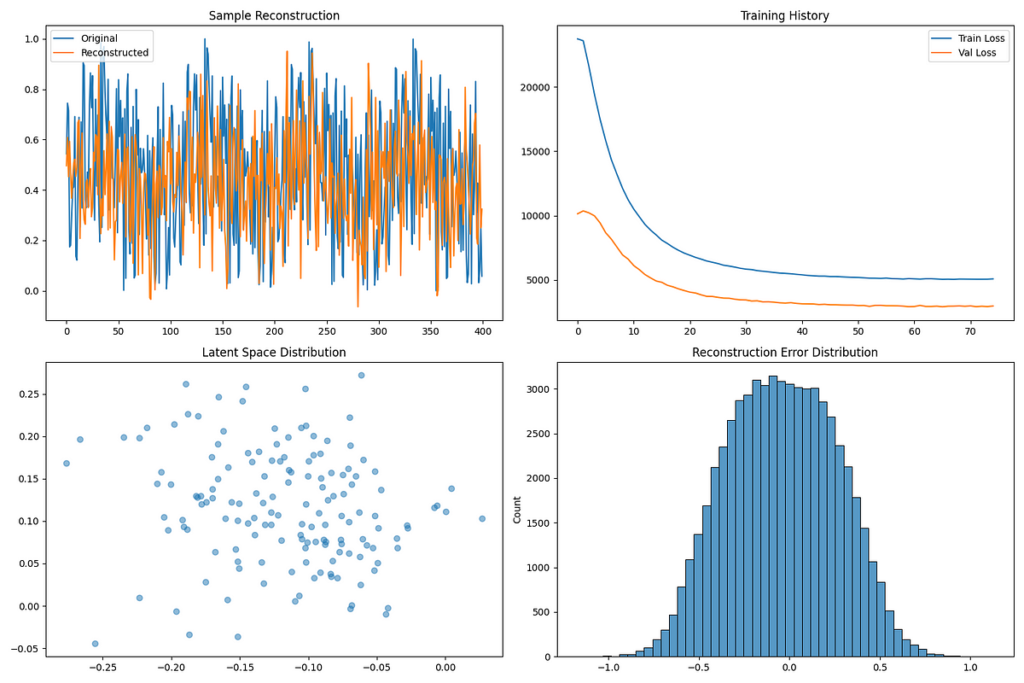
Helix Synth doesn’t simply predict — it creates. Utilizing a Variational Autoencoder (VAE), it generates solely new protein buildings:
- An encoder compresses protein sequences right into a 32-dimensional latent area (consider it as a compact “protein blueprint”).
- A decoder reconstructs these into full 3D tertiary buildings.
The VAE produced 5,003 artificial proteins with 90% confidence and a disentanglement rating of 0.9024 (a measure of how properly it separates distinct protein options). Nevertheless, the reconstruction error was 278.3618, suggesting room for refinement.
To make artificial proteins extra correct, Helix Synth makes use of a diffusion mannequin impressed by Denoising Diffusion Probabilistic Fashions (DDPM). This step refines 3D folds, making certain the generated buildings are biologically reasonable and practical.